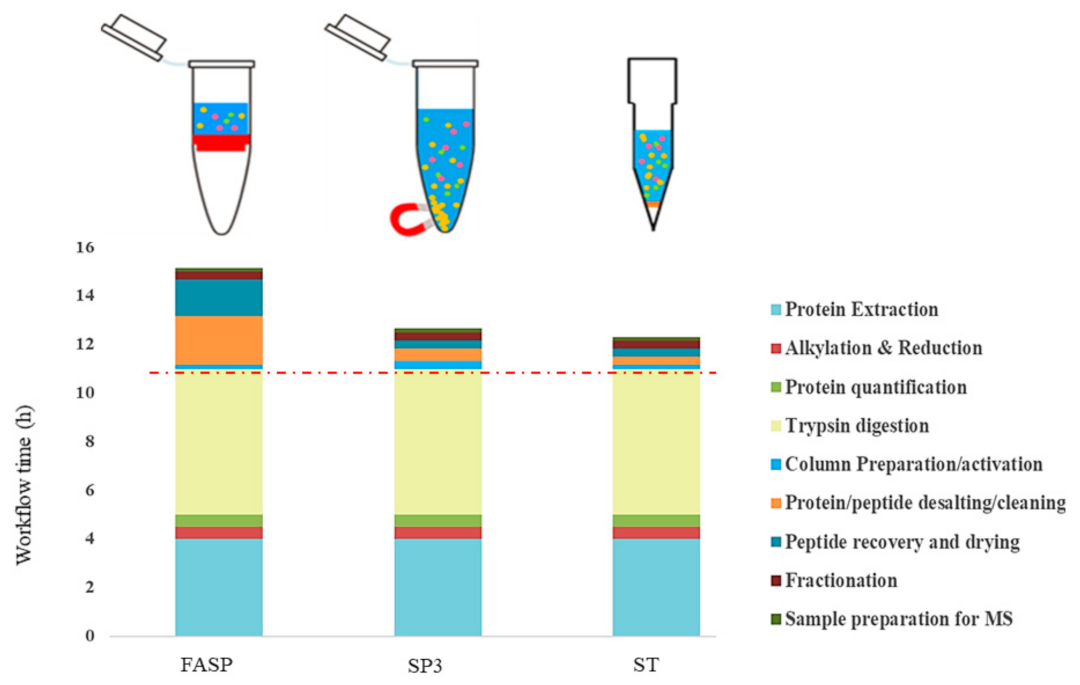
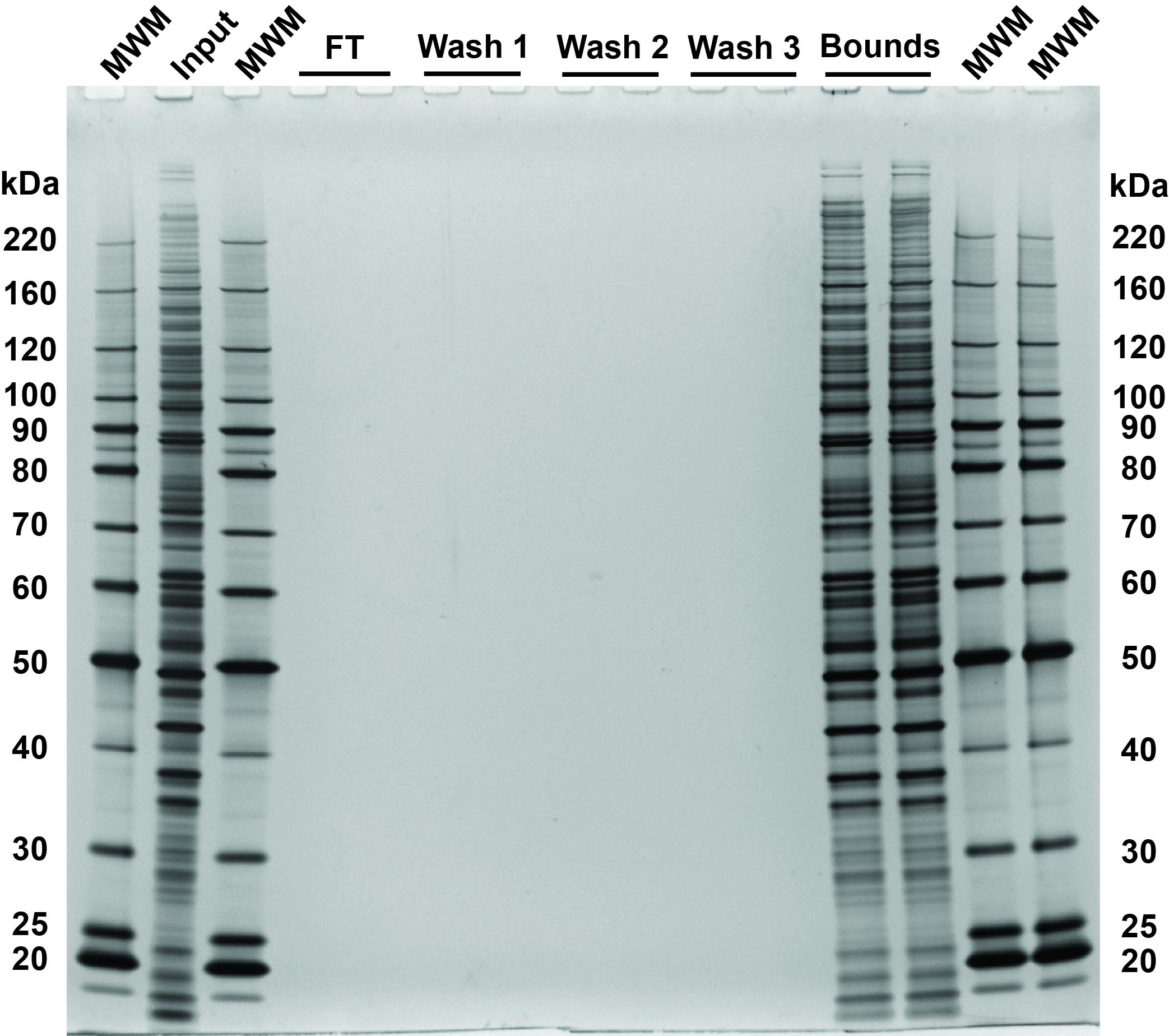
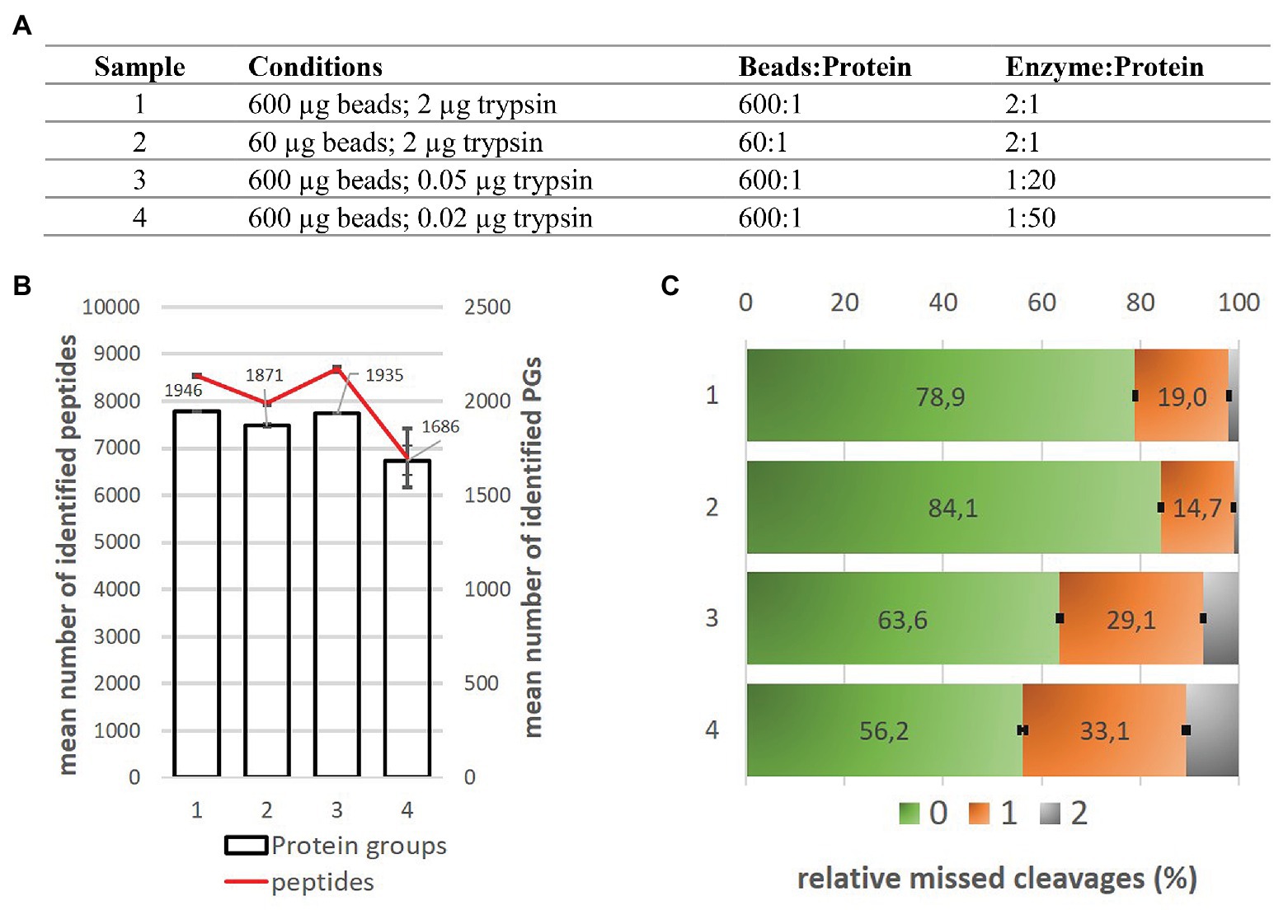
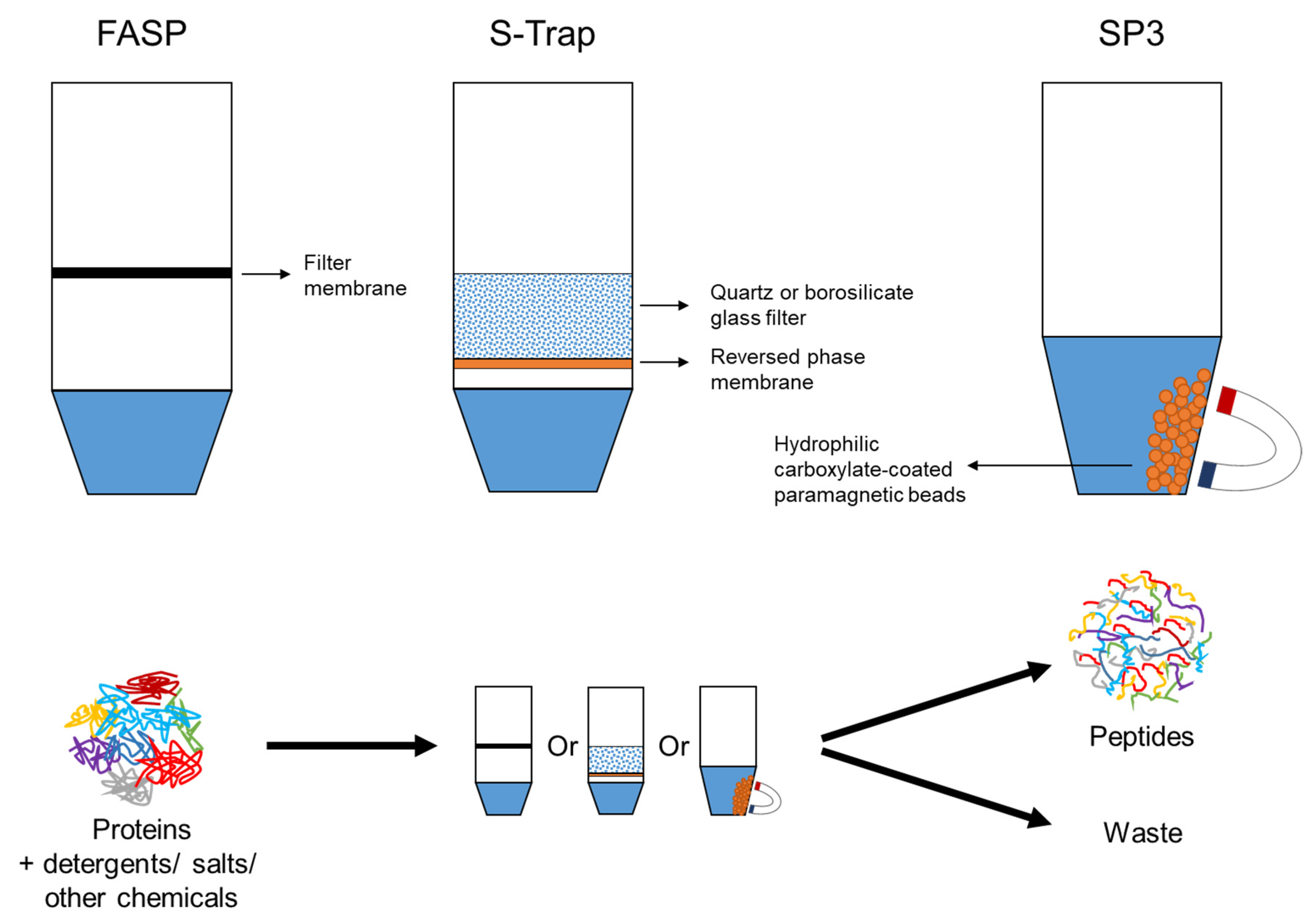
Multiplexed proteome profiling of carbon source perturbations in two yeast species with SL-SP3-TMT - ScienceDirect
Solvent Precipitation SP3 (SP4) enhances recovery for proteomics sample preparation without magnetic beads

Rahul Samant on X: "Deep proteome profiling—TMTsixplex, Lys-C+trypsin digestion, followed by off-line peptide fractionation into 62 fractions—showed almost identical results for SP3, SP4 (bead-free), and SP4 (+glass beads), WITH ONE NOTABLE EXCEPTION…

Frontiers | Evaluation of Sample Preparation Methods for Fast Proteotyping of Microorganisms by Tandem Mass Spectrometry

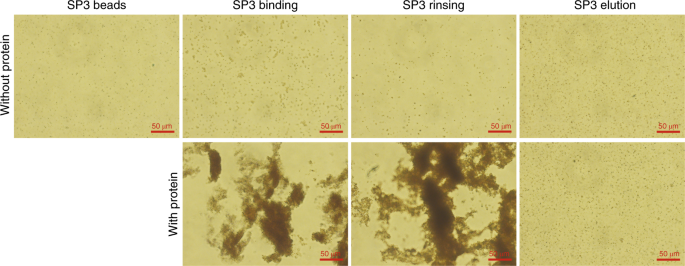
Extending the Compatibility of the SP3 Paramagnetic Bead Processing Approach for Proteomics | Journal of Proteome Research

Extending the Compatibility of the SP3 Paramagnetic Bead Processing Approach for Proteomics | Journal of Proteome Research

Solvent Precipitation SP3 (SP4) Enhances Recovery for Proteomics Sample Preparation without Magnetic Beads | Analytical Chemistry

Improving Proteome Coverage for Small Sample Amounts: An Advanced Method for Proteomics Approaches with Low Bacterial Cell Numbers - Blankenburg - 2019 - PROTEOMICS - Wiley Online Library

SP3-Enabled Rapid and High Coverage Chemoproteomic Identification of Cell-State–Dependent Redox-Sensitive Cysteines - ScienceDirect

Solvent Precipitation SP3 (SP4) Enhances Recovery for Proteomics Sample Preparation without Magnetic Beads | Analytical Chemistry

Implementing solid-phase-enhanced sample preparation for Co-Immunoprecipitation with Mass Spectrometry for C. elegans | bioRxiv

Solvent Precipitation SP3 (SP4) enhances recovery for proteomics sample preparation without magnetic beads | bioRxiv
Solvent Precipitation SP3 (SP4) enhances recovery for proteomics sample preparation without magnetic beads